All-Ions Deconvolution Software
In the omics-fields (e.g., proteomics, lipidomics, metabolomics, and exposomics) as many molecules as possible must be identified, often without prior knowledge. To annotate chemicals using liquid chromatography high-resolution mass spectrometry often molecular fragmentation is required (MS/MS). One approach to acquire MS/MS for every ion is to use data-independent analysis (DIA: e.g., all-ions and Q-RAI) where the entire m/z region of interest is selected for fragmentation simultaneously or in segments. DIA is also useful for quantitation on the fragment ion which often gives more selectivity, specificity, and sensitivity than on the precursor ion. For both applications we introduce IonDecon, a rapid, open-source software, which intakes mzXML files and exports deconvoluted fragmentation spectra in .ms2 format, reconstructing precursor/fragment relationships. The software can be used in tandem with FluoroMatch or LipidMatch to include all-ions for MS/MS based annotation, just simply place resulting .ms2 files with proper naming conventions in your input folder. View the Sewage Sludge (SRM 2781) analysis to see an example using IonDecon and FluoroMatch.
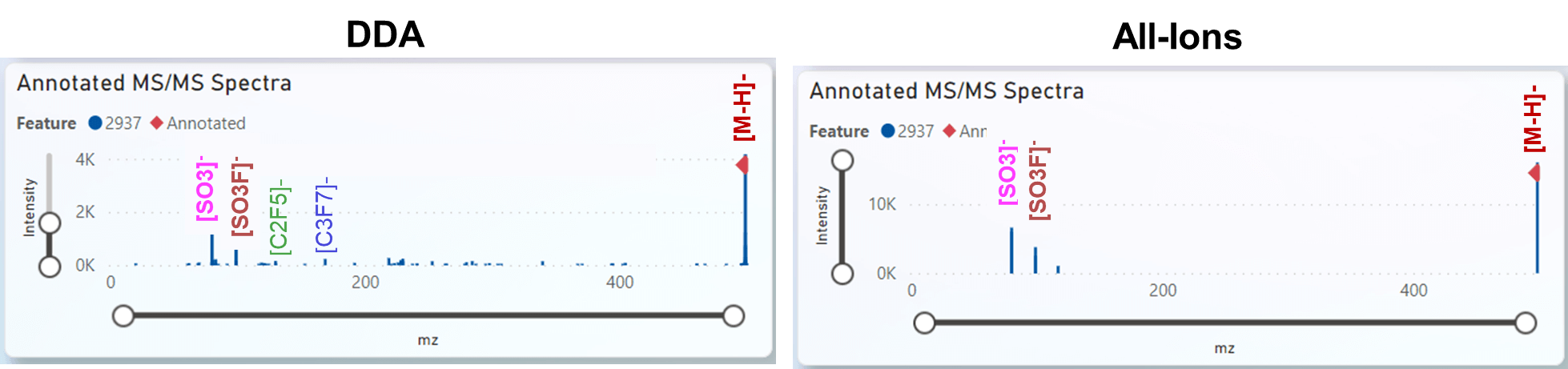
Above is a comparison of a MS/MS spectra from DDA and All-Ions after IonDecon deconvolution. Generally All-Ions covered more molecules then DDA, reducing false negatives, while only slightly increasing false positives.
This work is funded in part by Agilent Technologies, and software was designed by Jeremy Koelmel (Researcher, Krystal Pollitt Lab, Yale University), and written and co-designed by Michael Kummar and Nandini Abril.
Subscribe
0 Comments
Oldest
0 Comments
